Cheng-Yu Hou
Bioinformatician
10+ years work experience in Bioinformatics.
Advantages:
Experiences of both hands on Bioinformatics analysis and molecular biology experiments, which helps me to solve biology problems and to interpret questions as a biologist.
Phone: +886972105534 Line ID: Irous Mail: [email protected]

Education
National Taiwan Ocean University, Taiwan, Master degree, Bioscience and Biotechnology
2009 ~ 2011 | Thesis: Production of Chitooligosaccharides by Enzyme Engineering
Chinese Culture University, Taiwan, Bachelor degree, Life Science
Work Experience (10+ years in Bioinformatics)
NGS team, Microbio Group, Taiwan, Senior assistant reseacher, Dec 2021 ~ Current
1. Microbiota analysis(Gut, Skin)
2. Server and Database management
3. Statical analysis for biology data(microbiota, new drug development)
4. Data intergration, visualization and interpretation
BioInformatics team, Sofiva Genomics, Taiwan, Bioinformatician, Jun 2016 ~ Nov 2021
1. To intergrade demands from different teams in the lab and design (web/stand-along) tools with Graphical User Interface to generate data for reports.
2. Development of customized programs and flexible pipe-lines (Linux, Windows)
3. Analysis of disease-related NGS data (inherited disease, tumor)
4. Consulting for software and web-tools for NGS data analysis
Agricultural Biotechnology Research Center, Academia Sinica, Taiwan, Research Assistant, Jul 2012 ~ May 2016
1. Development of customized programs for NGS data filtering, retrieval, manipulation and mining
2. RNA Degradome data analyses
3. Computer maintenance
Publications
W.C. Lee#, B.H. Hou#, C.Y. Hou#, S.M. Tsao, Ping. Kao, H.M. Chen*(2020) Widespread EJC footprints in the RNA degradome mark mRNA degradation before the steady-state translation. Plant Cell.
Impact factor: 8.6, ranking: 4/453 in Plant Sciences
# These authors contributed equally to this work.
C.Y. Hou, W.C. Lee, H.C. Chou, A.P. Chen, S.J. Chou, H.M. Chen* (2016) Global Analysis of Truncated RNA Ends Reveals New Insights into Ribosome Stalling in Plants, Plant Cell.
Impact factor: 8.5, ranking: 4/209 in Plant Sciences
C.Y. Hou, M.T. Wu, S.H. Lu, H.M. Chen* (2014) Beyond cleaved small RNA targets: unraveling the complexity of plant RNA degradome data, BMC Genomics, 15(1), 15
Skills
NGS data analyses
- Variant calling: Human
- Whole exon sequencing : Human
- Barcode De-multiplex
- miRNA analysis : Arabidopsis, Rice
- RNA Degradome analysis : Yeast, Arabidopsis, Rice, Soybean, Grape, Maize, Human
- Ribo-seq analysis : Arabidopsis
Bioinformatics tools
- Read aligner: Bowtie, BWA, STAR
- Adaptor trimming tool: Trimmomatic, Fastx
- Genome Analysis Toolkit: GATK
- miRNA target prediction tool: psRNATarget, CleaveLand
- Genome browser: IGV, GenomeBrowse
- Powerful platform for microbiota analysis: Qiime2
- Motif-based sequence analysis tool: MEME suite
- RNA structure prediction tool: Vienna RNA
- Protein 3D structure prediction tool: (PS)2 server
Programming skills
- Perl
- Python
- Html, PHP
- MySQL(MariaDB)
- Visual Basic
- VBA
Molecular Biology skills
- Protein 3D structure prediction and functional engineering
- Gene cloning and protein expression
- DNA / Protein extraction, purification and gel electrophoresis
Statistics / Visualization tools
- Sigmaplot
- R package
Computer graphics tools
- CorelDRAW
- Photoshop
International Conferences
9th AYRCOB(Asian Young Researchers Conference on Computational and Omics Biology), Biopolis, Singapore
21.Feb.2016 - 22.Feb.2016 (supported by Academia Sinica)
Poster presentation: Footprints of ribosomes and exon junction complexes are widespread in RNA degradome
Computational RNA Biology conference, Cambridge, England
11.Nov.2014 - 13.Nov.2014 (supported by Academia Sinica RPAS travel award)
Poster presentation: Genome-wide analysis of truncated RNA ends reveals degradation of newly synthesized targets of nonsense-mediated mRNA decay and microRNA
7th AYRCOB (Asian Young Researchers Conference on Computational and Omics Biology) Tokyo, Japan
08.Sep.2013 - 10.Sep.2013 (supported by AYRCOB)
Selected oral presentation: Beyond the cleavage products of small regulatory RNAs: unraveling the complexity of plant RNA degradome data
Mandatory Military service Jun. 2011 – May 2012
Service in Artillery Company
About me
After I finished my mandatory military service in 2012, I entered Academia Sinica as a research assistant. I’m responsible for data mining and computer maintenance. Even though I don’t have much experience at the moment, I‘ve gained the necessary skills in a short period, such as program coding and designing customized pipelines to conduct biology research. My efforts were published on The Plant Cell and BMC Genomics.
After years of experience in Bioinformatics, I found I possess an excellent ability to gain new knowledge and apply it to my work. For instance, the SofivaPedigree in my portfolio, it's the first time that I'm applying python to my work, as you can see, SofivaPedigree is a python based stand-along CAD(Computer Aid Graphic Design) program, which is a piece of art for the first try. The ability to study independently must be my best gift to solve problems. Moreover, I found I possess high patience in data mining because I think to find what is meaningful in tons of data is very interesting, and feel satisfied by doing research. You will be satisfied working with a person who always loves to improve oneself’s ability.
Portfolio
SofivaPedigree
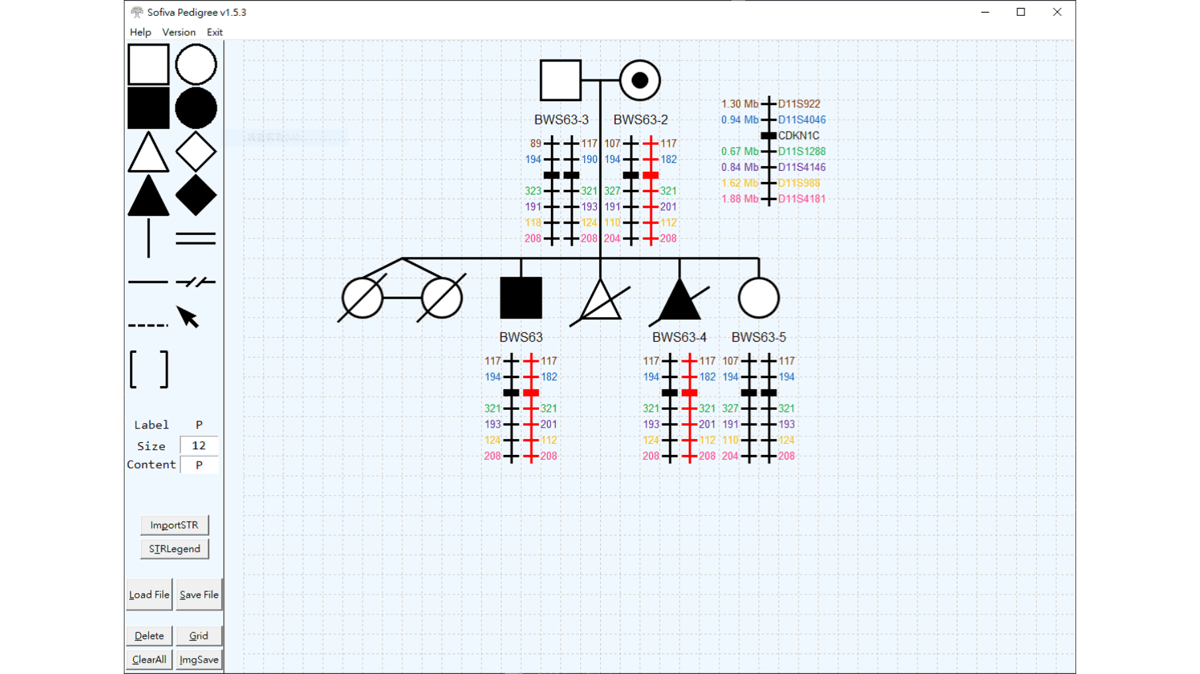
A python based stand-along CAD(Computer Aid Graphic Design) program. Designed for generating consistent outputs
Functions:
1. Save/Load file
2. Load setting file
3. Image output
4. Copy/Paste
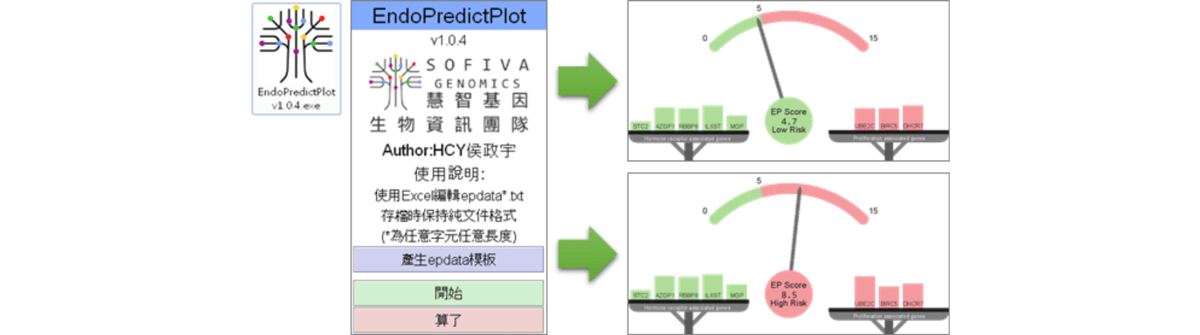
EndoPredictPlot
WiseAlignment
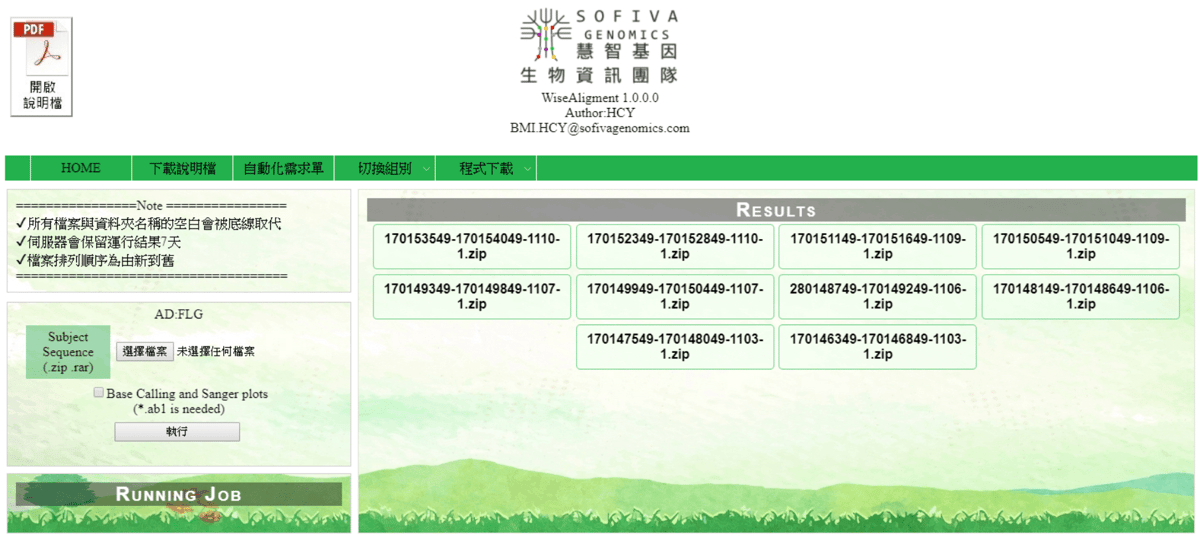
A web tool for sequence alignment.

Queue system
Showing queue, user can terminate their job manually.
WiseAlignment result
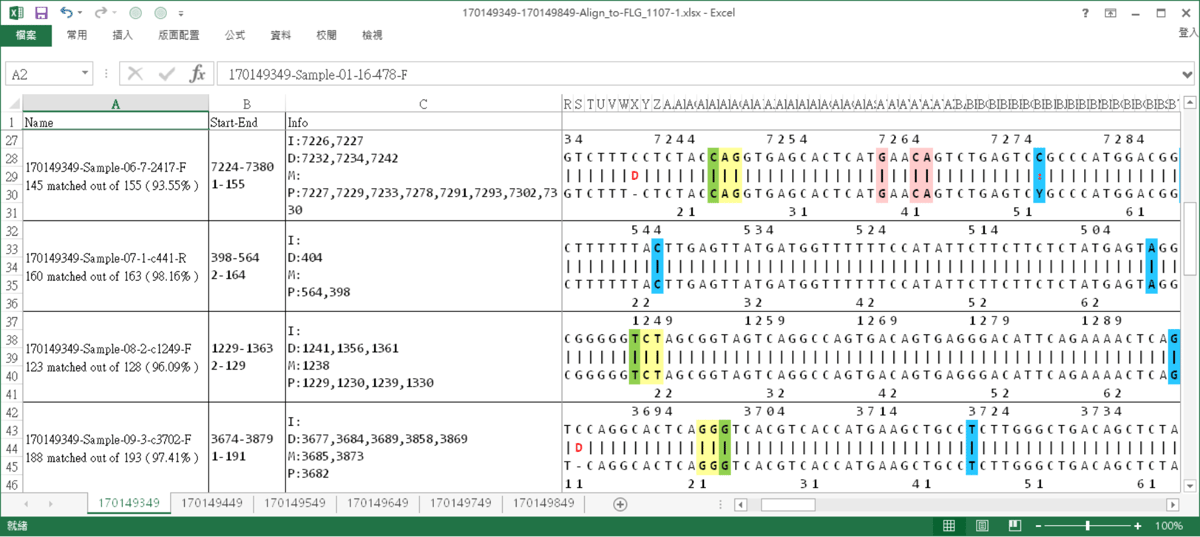
The results of alignment, the interested sites are marked.
AnnotationPlus

A web tool for searching annotation from multiple database.

Queue system
Showing queue, user can terminate their job manually.
AnnotationPlus result
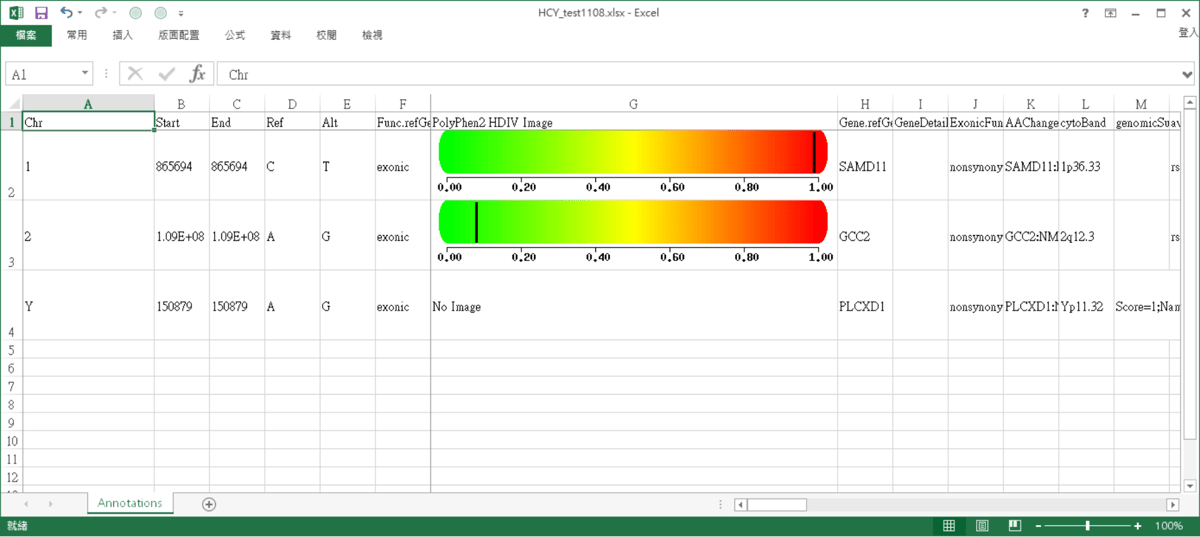
The results of annotation, shown in Excel.
Art work of computer graphics
Below is the art work plotted by CorelDraw and Photoshop
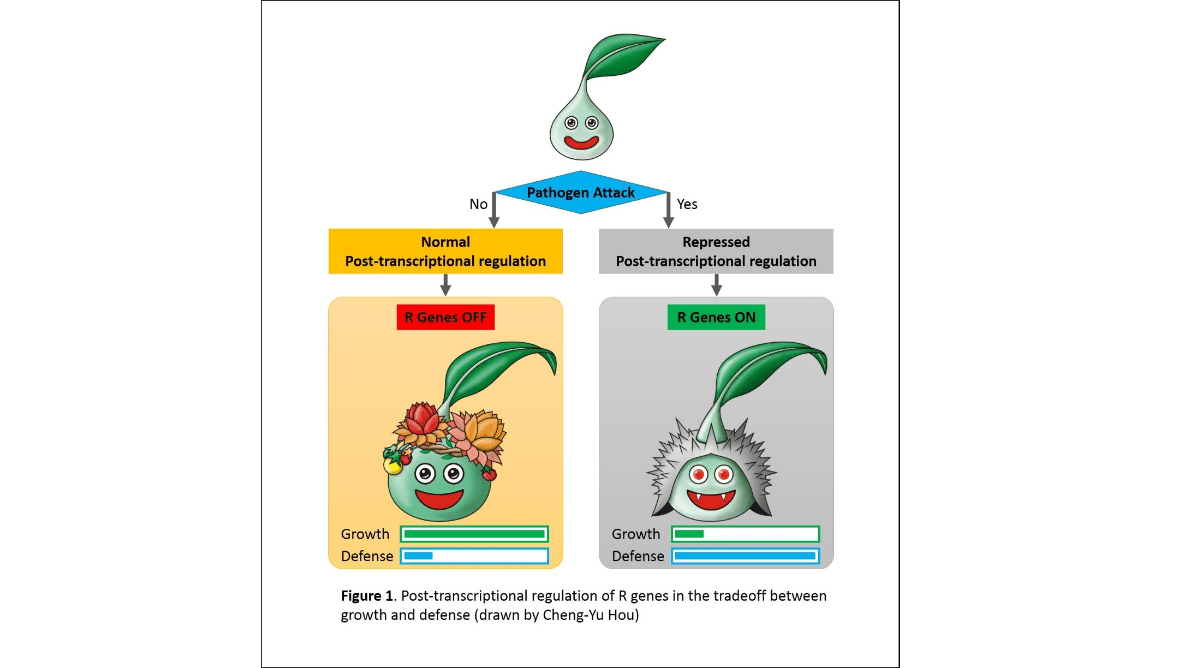
Plant defense mechanism
Illustrating a figure for explaining plants response differently in differ environments
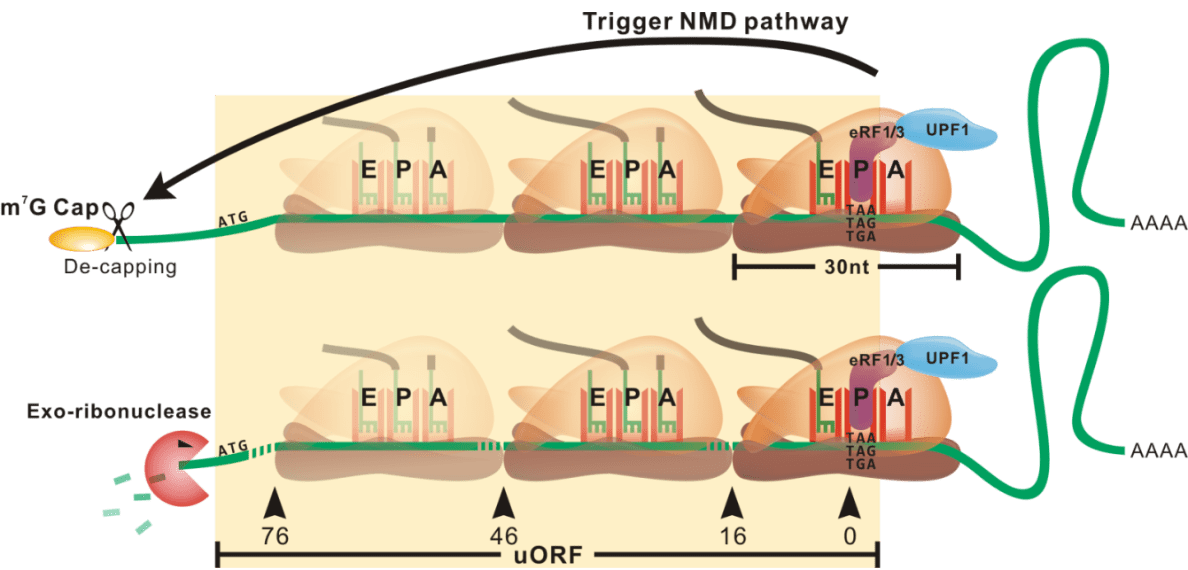
Model of ribosome stalling
Illustrating a biological model of ribosome stalling
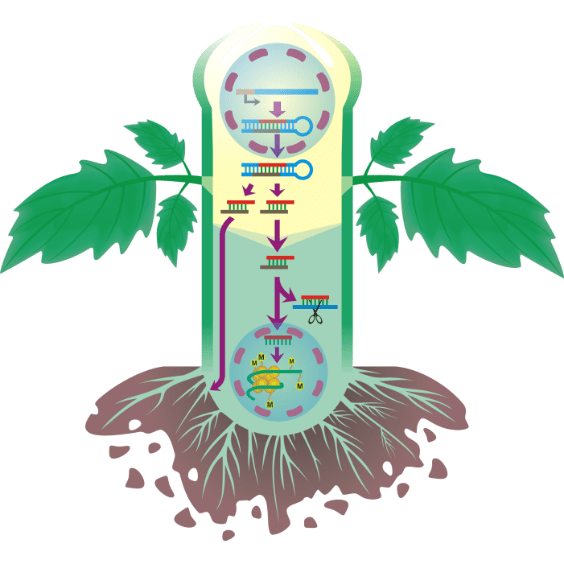
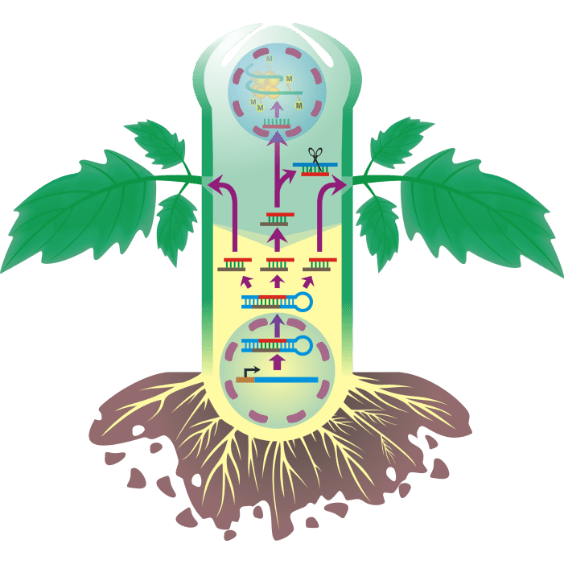
RNA movement
Illustrating figure of project for applying for funds explaining how RNAs interfere plant system
